Software
Phenonaut
 Data integration workflows for multiomics data take many forms across academia and industry. Efforts with limited resources often encountered in academia can easily fall short of data integration best practices for processing and combining high-content imaging, proteomics, metabolomics, and other omics data. We present Phenonaut, a Python software package designed to address the data workflow needs of migration, control, integration, and auditability in the application of literature and proprietary techniques for data source and structure agnostic workflow creation.
Data integration workflows for multiomics data take many forms across academia and industry. Efforts with limited resources often encountered in academia can easily fall short of data integration best practices for processing and combining high-content imaging, proteomics, metabolomics, and other omics data. We present Phenonaut, a Python software package designed to address the data workflow needs of migration, control, integration, and auditability in the application of literature and proprietary techniques for data source and structure agnostic workflow creation.
Reference: Steven Shave, John Dawson, Abdullah Athar, Cuong Nguyen, Richard Kasprowicz, Neil Carragher, “Phenonaut: multiomics data integration for phenotypic space exploration.” Bioinformatics, 2023.
PyBindingCurve
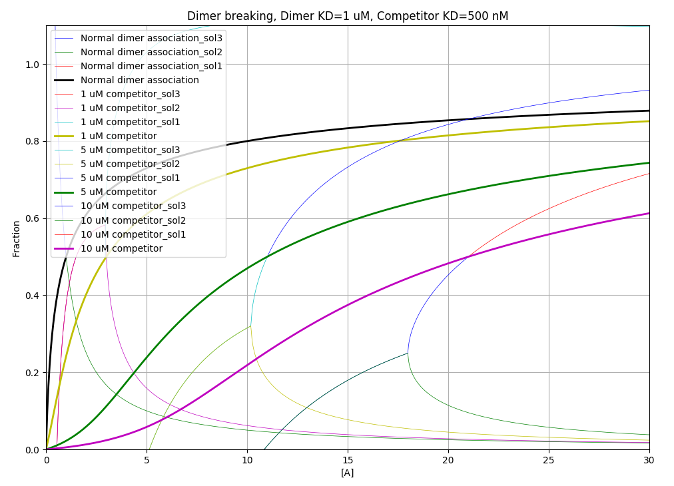 PyBindingCurve is a Python package for simulation, plotting and fitting of experimental parameters to protein-ligand binding systems at equilibrium. In simple terms, the most basic functionality allows simulation of a two species binding to each other as a function of their concentrations and the dissociation constant (KD) between the two species.
PyBindingCurve is a Python package for simulation, plotting and fitting of experimental parameters to protein-ligand binding systems at equilibrium. In simple terms, the most basic functionality allows simulation of a two species binding to each other as a function of their concentrations and the dissociation constant (KD) between the two species.
Reference: Steven Shave, Yan-Kai Chen, Nhan Pham, Manfred Auer, “PyBindingCurve, simulation, and curve fitting to complex binding systems at equilibrium.” Journal of Chemical Information and Modeling, 2021.
OpenFEPOPS
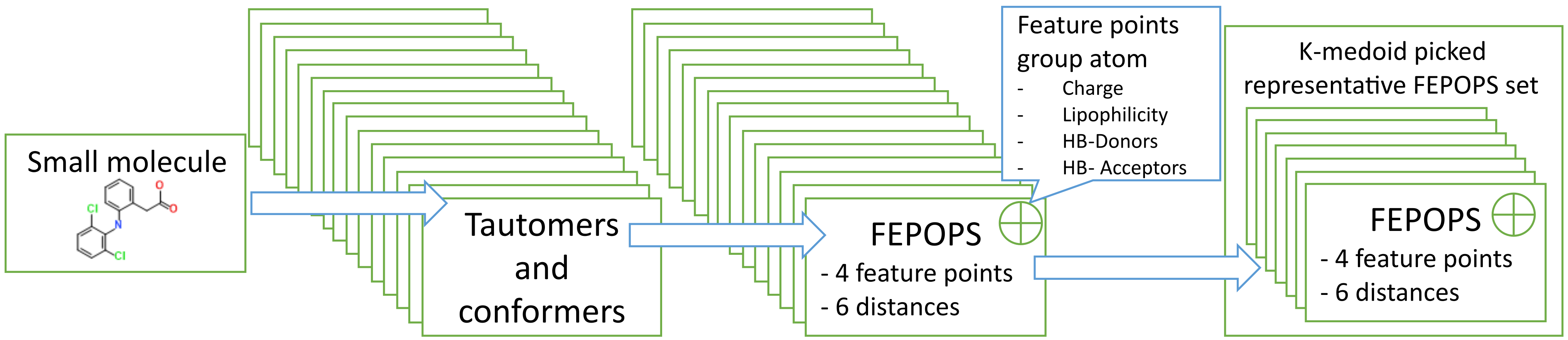 OpenFEPOPS is an open-source Python implementation of the FEature POint PharmacophoreS (FEPOPS) molecular similarity technique enabling descriptor generation, comparison, and ranking of molecules in virtual screening campaigns. Ligand based virtual screening is a fundamental approach undertaken to expand hit series or perform scaffold hopping whereby new chemistries and synthetic routes are made available in efforts to remove undesirable molecular properties and discover better starting points in the early stages of drug discovery. Typically, these techniques query hit molecules against proprietary, in-house, or publicly available repositories of small molecules in the hope of finding close matches which will display similar activities to the query based on the molecular similarity principle which states that similar molecules should have similar properties and make similar interactions.The central idea behind FEPOPS is reducing the complexity of molecules by merging of local atomic environments and atom properties into ‘feature points’. This compressed feature point representation has been used to great effect as noted in literature, helping researchers identify active and potentially therapeutically valuable small molecules. By default, OpenFEPOPS uses literature reported parameters which show good performance in retrieval of active lead- and drug-like small molecules within virtual screening campaigns, with feature points capturing charge, lipophilicity, and hydrogen bond acceptor and donor status.
OpenFEPOPS is an open-source Python implementation of the FEature POint PharmacophoreS (FEPOPS) molecular similarity technique enabling descriptor generation, comparison, and ranking of molecules in virtual screening campaigns. Ligand based virtual screening is a fundamental approach undertaken to expand hit series or perform scaffold hopping whereby new chemistries and synthetic routes are made available in efforts to remove undesirable molecular properties and discover better starting points in the early stages of drug discovery. Typically, these techniques query hit molecules against proprietary, in-house, or publicly available repositories of small molecules in the hope of finding close matches which will display similar activities to the query based on the molecular similarity principle which states that similar molecules should have similar properties and make similar interactions.The central idea behind FEPOPS is reducing the complexity of molecules by merging of local atomic environments and atom properties into ‘feature points’. This compressed feature point representation has been used to great effect as noted in literature, helping researchers identify active and potentially therapeutically valuable small molecules. By default, OpenFEPOPS uses literature reported parameters which show good performance in retrieval of active lead- and drug-like small molecules within virtual screening campaigns, with feature points capturing charge, lipophilicity, and hydrogen bond acceptor and donor status.
Reference: Chen, Y., Houston, D. R., Auer, M., & Shave, S. (2023). OpenFEPOPS: A Python implementation of the FEPOPS molecular similarity technique. Journal of Open Source Software, 8(91), 5763.
Smi2Svg
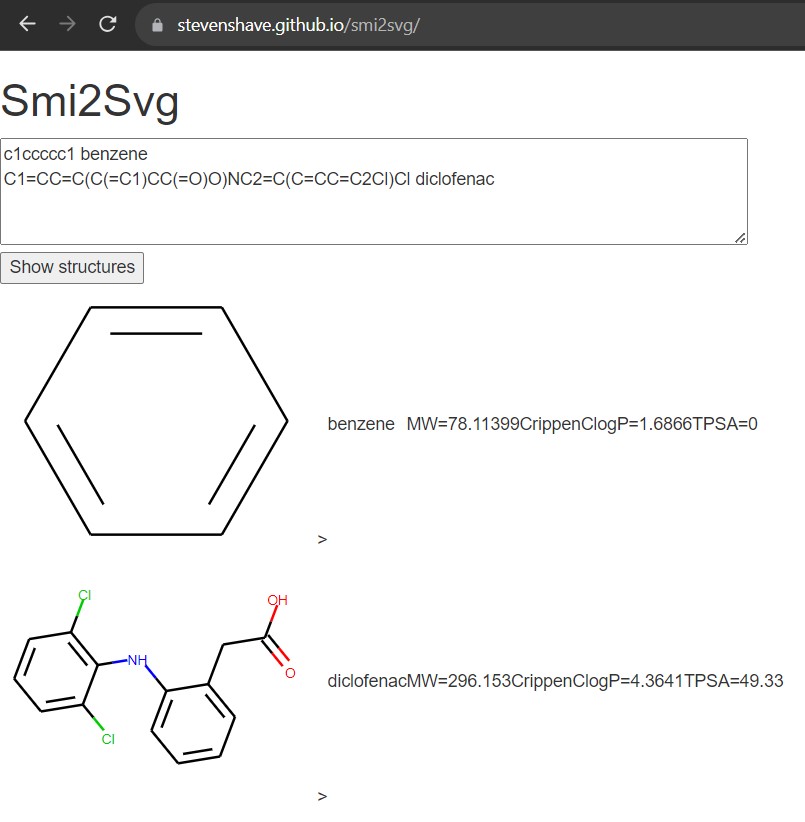
A useful little web tool I built using WASM compiled RDKit to quickly visualise molecules from their SMILES strings, runs completely in the browser.
https://stevenshave.github.io/smi2svg/




